Each of the tutorials builds on the preceding tutorials, and they should be completed in order.
This is the multi-page printable view of this section. Click here to print.
Tutorials
1 - Logging in
In this tutorial, we’ll go over the steps needed to log into HortaCloud and start the Horta application.
HortaCloud only
This tutorial applies to HortaCloud only.Prerequisites
- You should know the URL for the HortaCloud instance that you will be using.
- The administrator of the instance should have created an account for you.
- You should know the username and password for that account.
The screenshots below were taken from the Janelia instance in June 2022.
Steps
The steps to log in to HortaCloud and starting the Horta application are similar to the steps for many web applications. The procedure is straightforward, but we’ve provided screenshots of every step to help troubleshooting when things go wrong.
Navigate to your HortaCloud website’s URL in your browser.
You are probably already on the site, if you’re reading this documentation! Click the button on the right labeled “Login to HortaCloud”.
All major browsers are supported: Safari, Google Chrome, Mozilla Firefox, and Microsoft Edge.
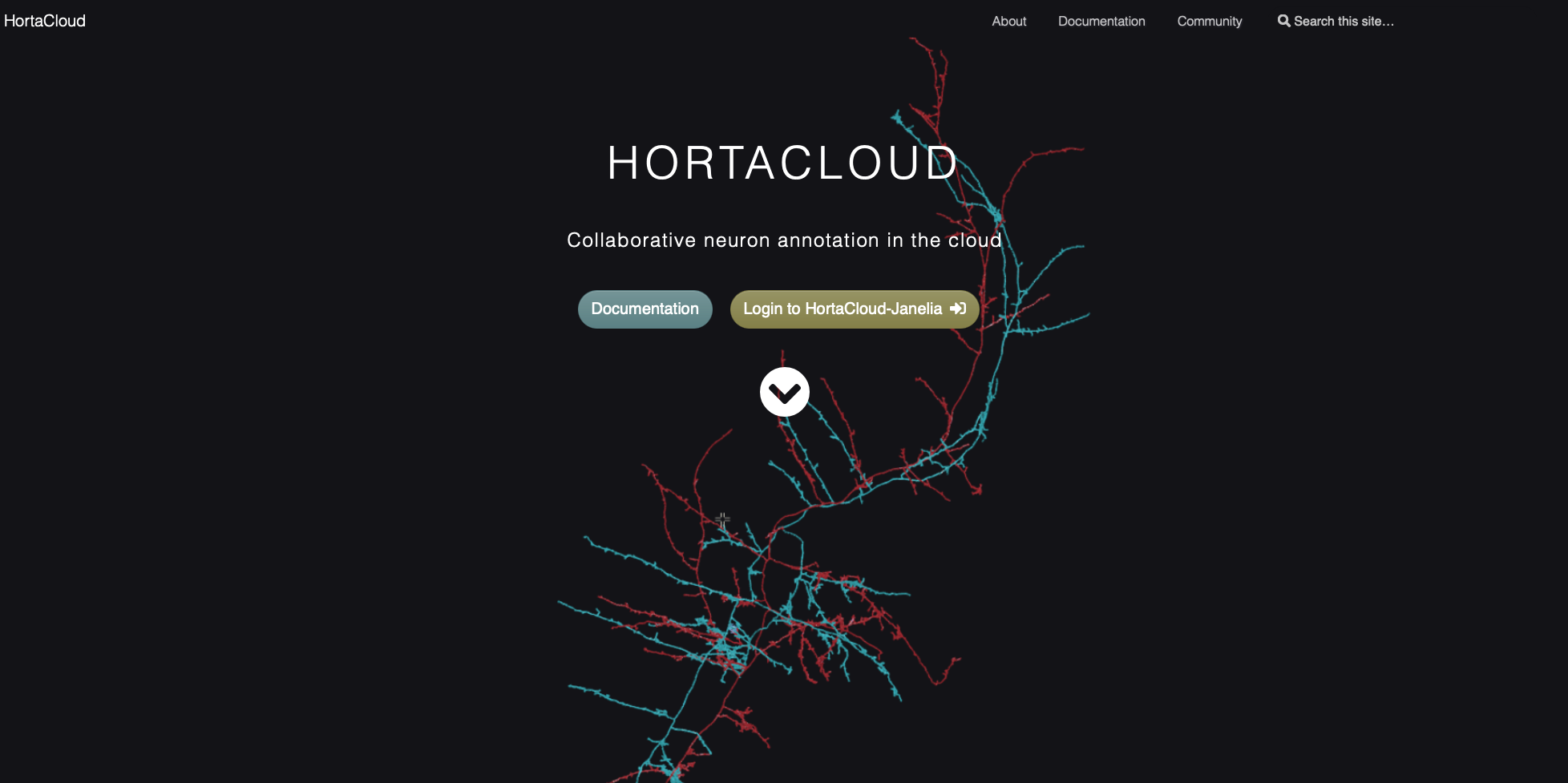
Enter your username and password given to you by your local administrator in the entry fields, then click the “Login” button.
On this page, there are also links for changing your password and for requesting a password reset if you have forgotten your password.
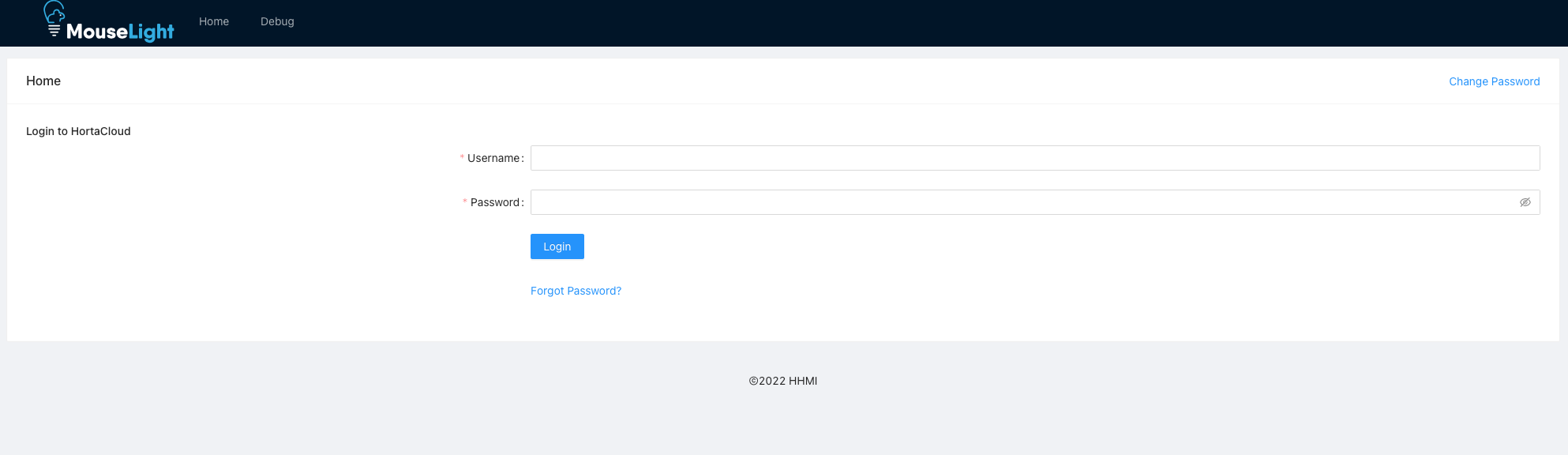
Click “AppStream Login” to continue.
This screen also contains a change password link, and if you are a Horta application administrator, you will also see tools for managing users and groups in Horta.
Click “Horta Workstation” to start the application.
This is currently the only application in HortaCloud.

Wait for application startup
As the application starts, which can take a couple minutes, you will see a wait screen (first image below). The percentage should steadily increase. After that, you will see a brief console screen (second image below), then the Horta application itself (third image below).
The list of folders you see in the “Data Explorer” in Horta will be different and will depend on what data is available in your instance and which data you personally have permissions to see.

Troubleshooting
In most cases, if you have trouble launching the Horta application, you should contact your local HortaCloud administrator. If the application launches but you do not see the expected data, you should contact the local Horta application administrator. Of course, these may be the same person.
Ending your session
You can end your session by clicking on the rightmost icon on the AppStream toolbar and choosing “End Session”.

2 - Viewing images
In this tutorial, we’ll go over the steps needed to load images in Horta and view them.
HortaCloud-focused
This tutorial is focused on HortaCloud. However, most of it applies to the desktop version of Horta as well. If you can launch Horta on the desktop, and you see data in the Data Explorer, you should be able to follow most of the other steps of the tutorial.Prerequisites
- You should be able to log into HortaCloud and launch the Horta application, as detailed in the previous tutorial.
- Your HortaCloud instance should have access to the Janelia MouseLight data on Amazon AWS S3.
- If not, you should have access to some similar data in your HortaCloud instance.
Steps
Start the Horta Application
Launch the Horta application. See the previous tutorial “Logging in” for more detail.
Locate and examine data in the Data Explorer
The “Data Explorer”, which appears by default in the upper left panel of Horta, shows a representation of all the data in Horta that you are allowed to see. This data is organized in folders, both created by the system and created by users. The exact list of folder you see will differ depending on which data you have permission to see.
Click on the triangle to the left of the folder named Home with the owner mouselight, which appears immediately to the right of the folder name, in yellow. This will open the folder to show its contents.
Click on the triangle to the left of the “3D Tile Microscope Samples” folder, also owned by mouselight. You will see a long list of samples, indicated by a green flask icon. Each of these sample represents a group of image files on disk or in an AWS S3 bucket.
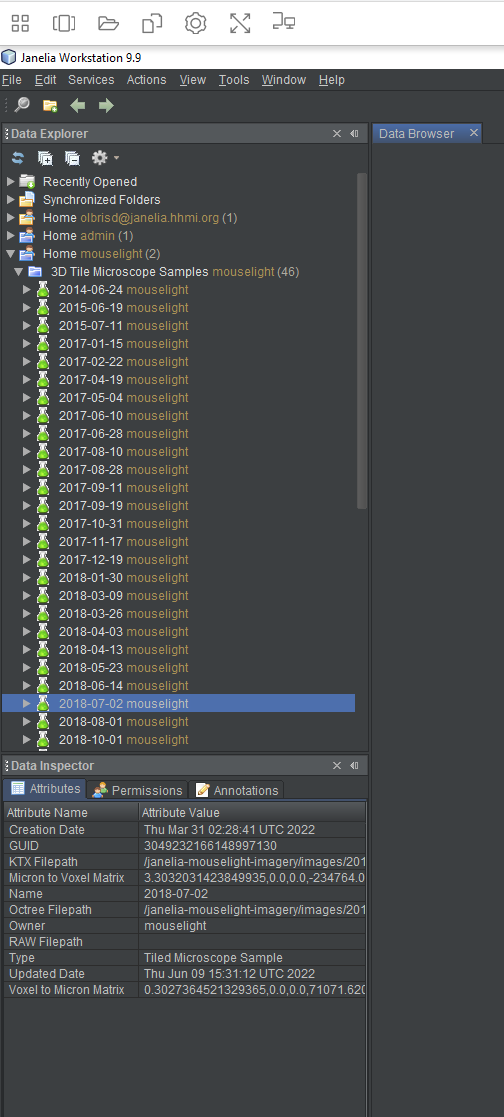
For this tutorial, we’re going to look at one of the Janelia Mouse Light samples, the 2018-07-02 sample. Scroll down the list of samples until you see it. Left-click on the sample to select it. You will see information appear in the “Data Inspector”, located just below the Data Explorer.
The three tabs of the Data Inspector each show information about the sample. The “Attributes” tab contains typical metadata like name, creation/modification date, and globally unique identifier (GUID). It also shows the sample’s owner and the location of the image files that belong to the sample. The “Permissions” tab shows which users or groups can read or write to the data. There’s also a button which allows someone to grant permissions to other users to see or edit the data. The third tab, “Annotations”, is not used by Horta and is only used with other tools in the desktop workstation client.
Open a sample in Horta
Now that we’ve located the 2018-07-02 sample, it’s time to open it in Horta.
Do one of two things:
- Right-click the sample, and choose “Open in Horta”.
- Left-click the sample to select it; then choose
Horta>Open in Hortafrom theActionsmenu.
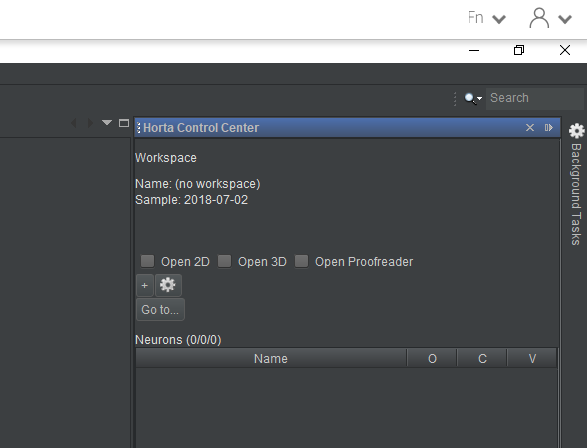
At this point, not a lot will visibly happen. When you open a sample in Horta, all that happens is that the dataset’s metadata is loaded into the application. The images themselves are not yet loaded. You will see that in the “Horta Control Center” panel at right, near the top, the “Sample” field will now show the name “2018-07-02”. That’s all (screenshot below). In the next tutorial (“Tracing neurons”), we’ll see that opening a workspace in Horta populates much more information in the UI. The “Concepts” section of the documentation has more information on the difference between samples and workspaces.
Open the Horta 3D window
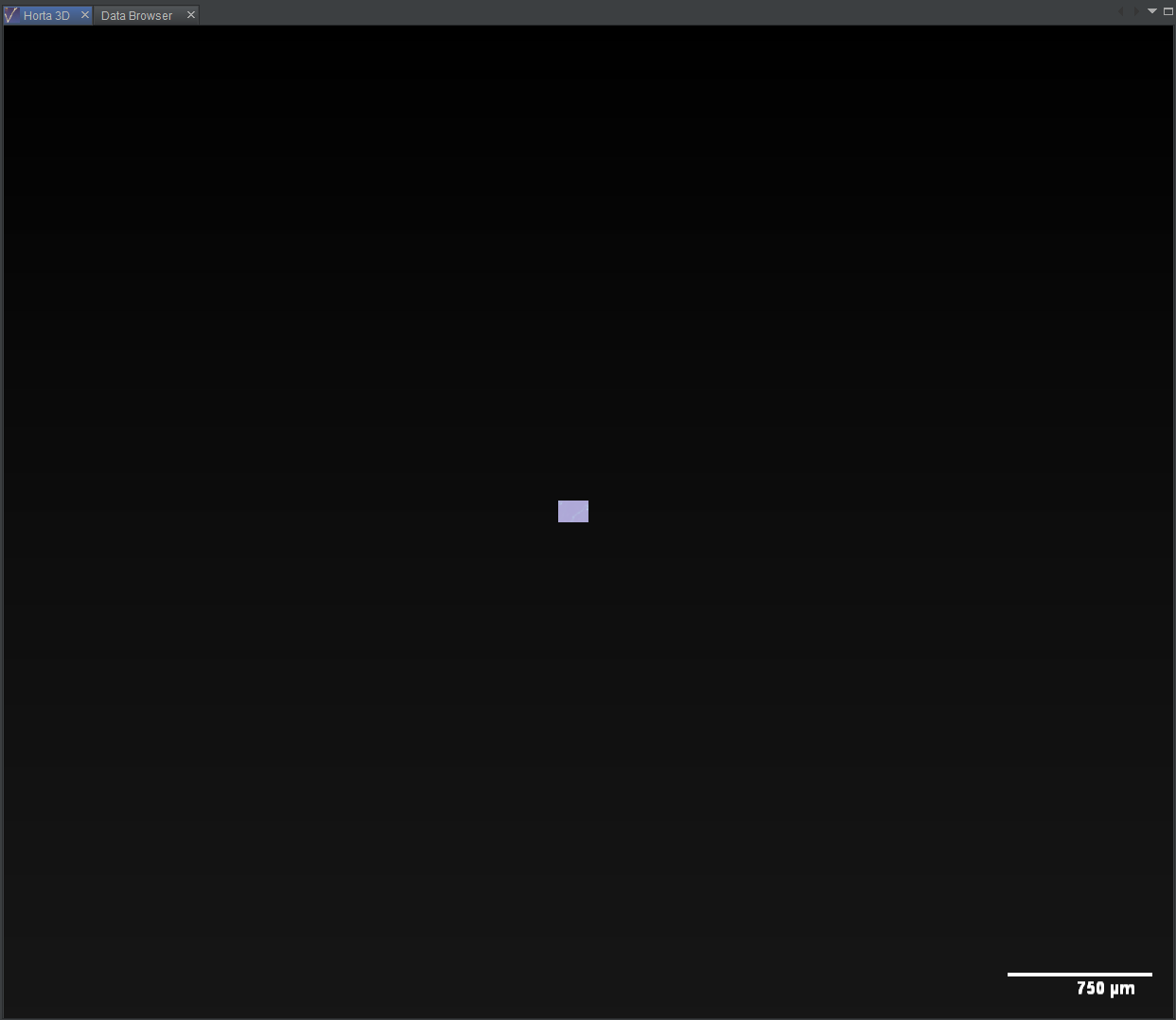
It’s time to finally look at some images. For this demonstration, we’ll start with the 3D images. Near the top of the Horta Control Center, find the checkbox labeled “Open 3D”. Click it so it’s checked.
You will see a window tab titled “Horta 3D” open in the center panel. The first 3D images will also load. Initially, it’ll be underwhelming. Probably you’ll see one small bright rectangle in the center of the screen. In the next section, we’ll see how to adjust the color. For now, if you left-click in the 3D window, more data will load to cover most of the window.
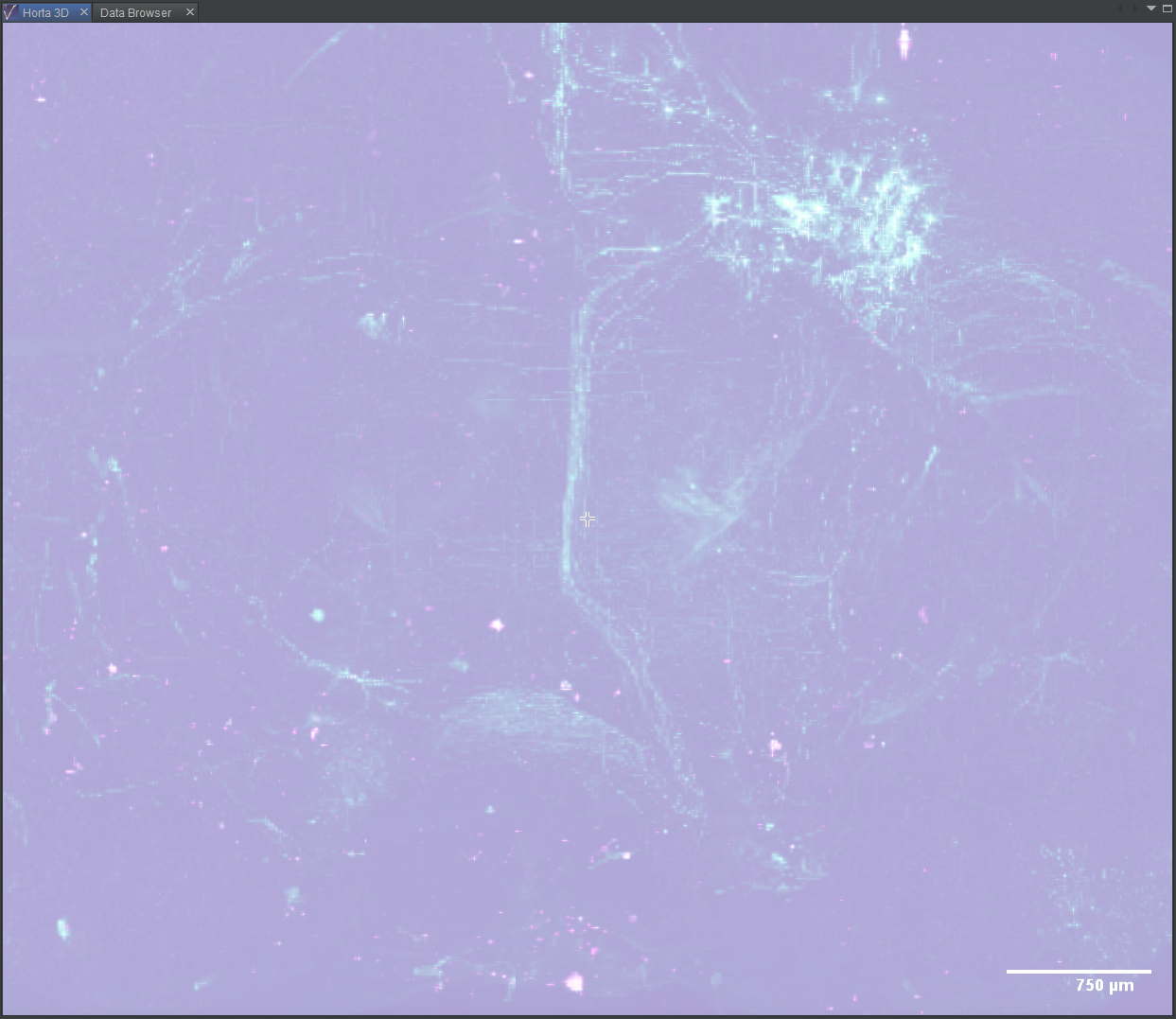
Explore the data in 3D
Adjust colors
The default color settings are not good; in general, the data will appear to be a washed out bright, light blue/purple. In order to adjust the color, first we need to go to the “Windows” menu and choose Horta > Color Sliders.
Often when the sliders are first opened, their panel will not be the correct height. You can adjust this. Like other panels in Horta, the color slider panel may be resized and docked or undocked from the main window as you like.
Also, when the color sliders are first opened, often the 3D image will be redrawn, or potentially not drawn at all. If this occurs, left-click in the 3D window to trigger a redraw.
Even though the data only has two channels (a signal channel and a reference channel), you’ll see three sliders. The third blue slider has a specialized use that we’re not going to cover in this tutorial. Click the eye next to the blue slider, and the blue channel will be hidden.
In practice, you will likely spend a lot of time tuning the color settings so the desired biological structures are most visible. For this tutorial, we’ll cover a few simple steps to make the data more visible:
- Click the eye icon to the left of the top, green slider. This turns the green channel data invisible.
- Now drag the leftmost slider of the middle, purple slider to the right. Do this until the blocky, purple background of the image tiles disappears and you can see the purple outline of the brain. For this data, you’ll be dragging the slider until it’s midway between the left edge and the center lock icon below the sliders.
- Now do it again for green. Click the eye next to the green slider to show the green channel, and the eye next to the purple channel to hide it. Drag the leftmost slide of the green bar until, again, the blocky background disappears and the brain outline is visible. For this data, try setting it a bit to the left of the middle purple slider.
- Click the eye next to the purple slide so both data channels are visible again.
At this point (screenshot below), the labeled neurons, though, stand out in green against the purple background. Feel free to experiment with the other sliders. In practice, some tracers prefer to adjust the settings until the background is nearly invisible, so that the neuron signal is prominent, even if dim.
Note: Color settings are not saved for samples! In the next tutorial, we’ll see how these settings are saved in workspaces.
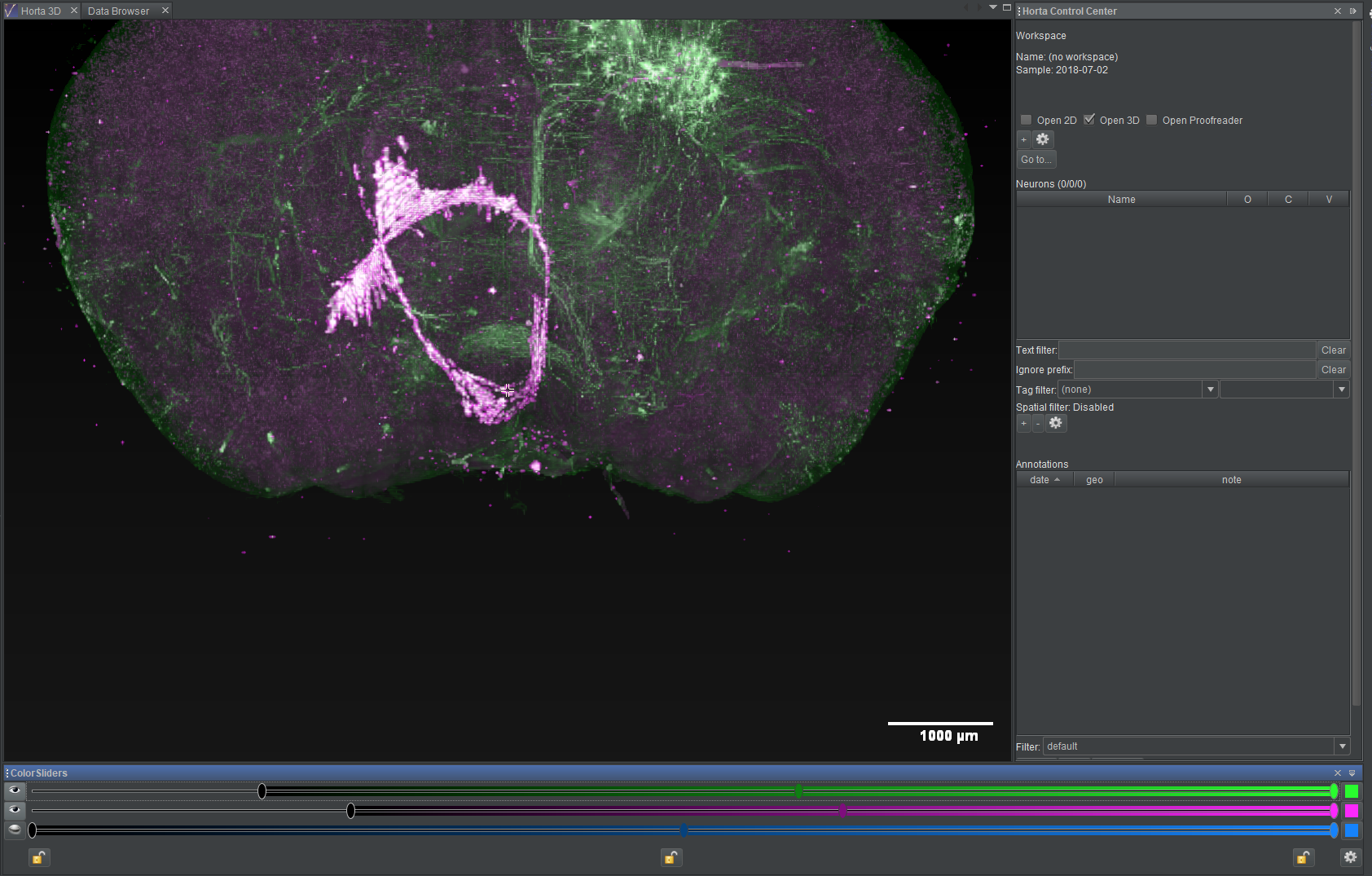
Pan, zoom, rotate
Navigating the 3D data is done using the mouse.
To navigate in space (pan):
- left-click to center on a location
- left-drag to move the image in the view plane
- as you navigate, images will be loaded and unloaded
To zoom in and out:
- use the scroll wheel to zoom in or out; as you zoom, higher or lower resolutions images will be loaded
- note that as you zoom in, you will see less and less of the thickness of the brain; that is, the displayed data will come from a projection depth that is smaller, allowing you to more clearly isolate smaller structures as you zoom in
To rotate in 3D:
- middle-click and drag, or hold down the shift key and left-click and drag; the cursor will change to two curved arrows, and the image will rotate in three dimensions
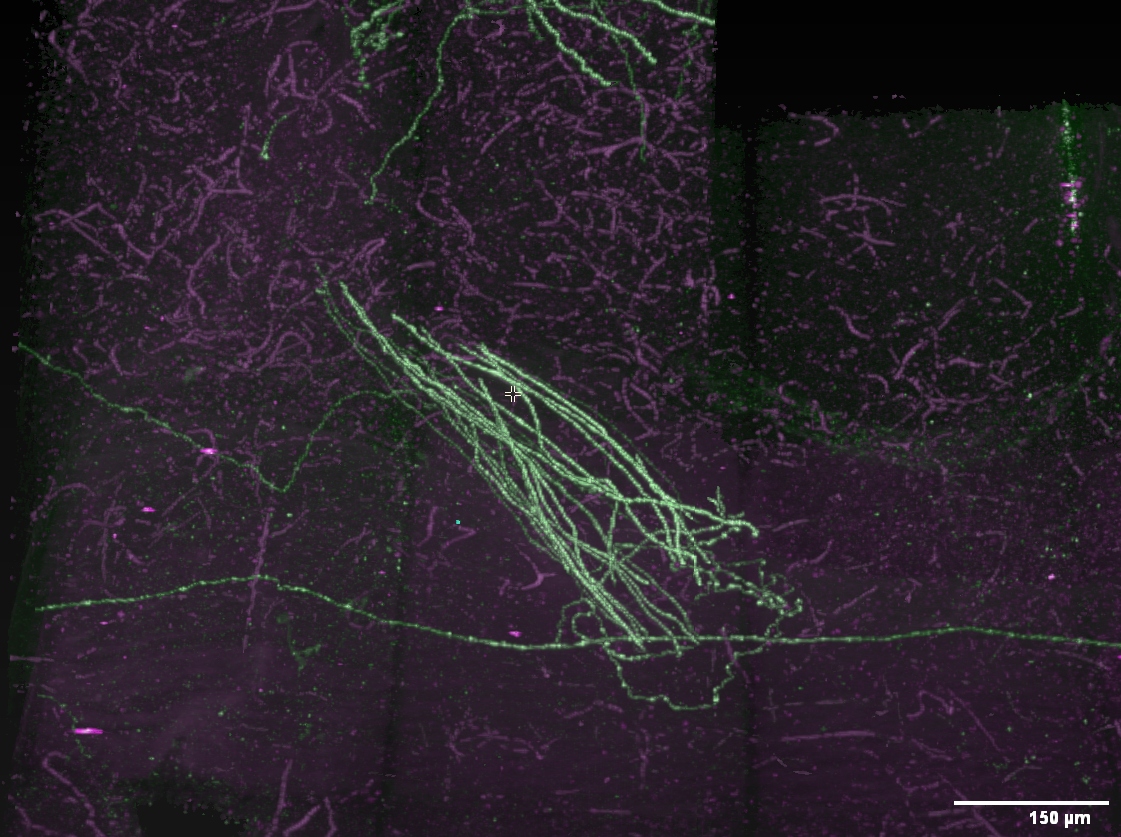
Open the Horta 2D window
HortaCloud
Note that HortaCloud does not store high-resolution 2D images! When you zoom in, the images will be blurry.
In the Horta desktop application, when you zoom in, higher-resolution images will automatically be loaded.
Loading the 2D images is done just like for 3D. Near the top of the Horta Control Center, find the checkbox labeled “Open 2D”. Click it so it’s checked.
You will see a window tab titled “Horta 2D” open in the center panel. The first 2D images will also load.
Explore the data in 2D
Adjust colors
As with 3D, the default color settings are almost never useful, and we’ll quickly adjust them to make the data more visible. In practice, you will likely spend a lot of time tuning the color settings so the desired biological structures are most visible.
The colors of the two data channels are controlled by the sliders and buttons at the bottom of the middle panel in the “Horta 2D” tab. They are distinct from the sliders used to adjust the 3D colors.
NOTE: Sometimes the color slider do not draw correctly when the sample initially opens in 2D. If this is the case, you will see two small checkboxes and two small plus signs at the right of the empty area below the 2D image. Toggle either on of the check boxes off and on, and the sliders should redraw.
Here are the steps to make some minimal adjustments to the colors:
- Click the eye icon to the left of the top, green slider. This turns the green channel data invisible.
- Now drag the leftmost slider of the lower, purple slider to the right. Do this until the blocky, purple background of the image tiles disappears and you can see the purple outline of the brain. For this data, you’ll be dragging the slider until it’s midway between the left edge and the center lock icon below the sliders.
- Now do it again for green. Click the eye next to the green slider to show the green channel, and the eye next to the purple channel to hide it. Drag the leftmost slide of the green bar until, again, the blocky background disappears and the brain outline is visible. For this data, try a location just to the left of the middle purple slider.
- Click the eye next to the purple slide so both data channels are visible again.
At this point (screenshot), the labeled neurons stand out in green against the purple background. Feel free to experiment with the other sliders. In practice, some tracers prefer to adjust the settings until the background is nearly invisible so the neuron signal is prominent, even if dim.
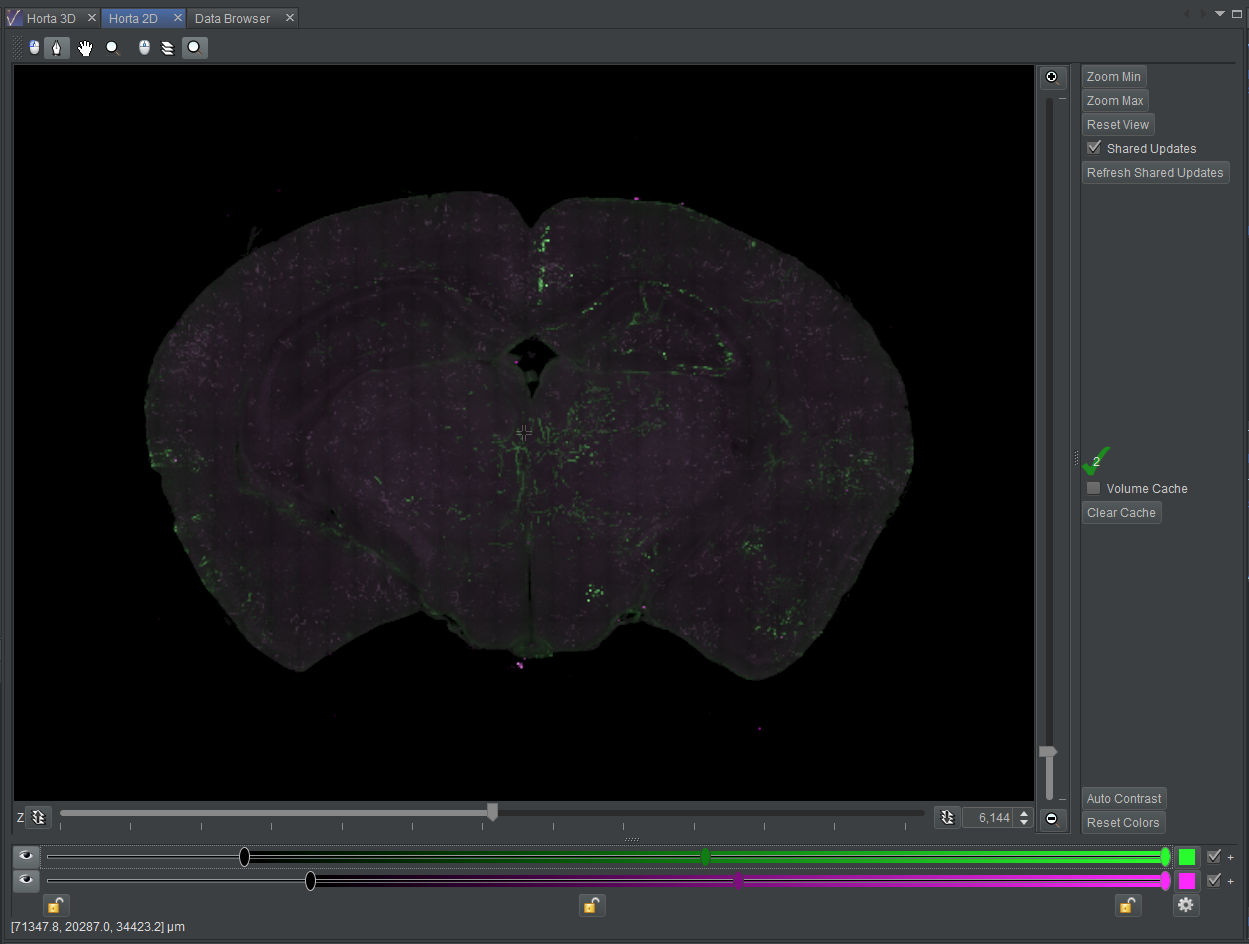
Note: Color settings are not saved for samples! In the next tutorial, we’ll see how these settings are saved in workspaces.
Pan, scroll through z, zoom
Navigating the 2D data is done using the mouse.
To navigate in the x-y plane, you can:
- double-left-click on any location to center the view on that point
- middle-click and drag to pan the image to any location
To navigate along the z-axis, you can:
- use the scroll wheel to change the visible plane
- drag the horizontal slider below the 2D view to change the visible plane
- change the plane number visible in the plane number box to the right of the plane slider by either typing a plane number in the box, or by clicking the up and down arrows in the box
To zoom the image:
- hold down the shift key and spin the scroll wheel
- drag the vertical slider to the right of the 2D view
- remember, HortaCloud does not has high-resolution 2D images; when you zoom in, it will be blurry!
x, y, z locations
Status bars
As you move the mouse cursor in either the 2D or 3D windows, you will see the x, y, z location of the cursor displayed in the status bars.
- in 3D, the status bar is at the very bottom of the Horta application window
- in 2D, the status bar is at the bottom of the 2D window; the 3D status bar in this case is not accurate, and it does not update
- in both cases, the
x, y, zlocation is given in microns (µm), as a floating-point number
The screenshot below shows both status bars, with the 3D bar at the bottom and the 2D bar above it.
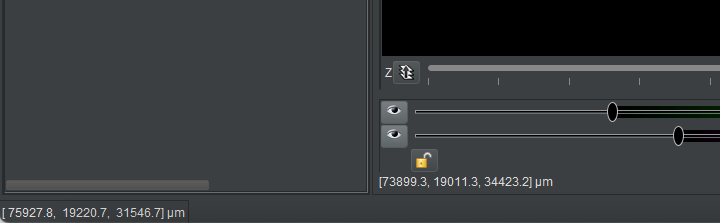
Copying location values
You can copy the the current x, y, z location to the clipboard by right-clicking in the view.
- in 3D, right-click and choose “Copy Micron Location to Clipboard”
- in 2D, right-click and choose “Copy Micron Coords to Clipboard”
- in both cases, the copied text has the following form:
[68665.88,48154.89,26670.117]- this is convenient for pasting into eg Matlab or Python
- (HortaCloud) see the “AppStream Basics” section in this documentation for how to transfer the data from AppStream’s clipboard to your local system’s clipboard
Go to point
If you know the x, y, z location you’d like to navigate to, and that data is on the clipboard, click the “Go to…” button that appears just above the neuron list in the Horta Control Center. Paste in the data from the clipboard and click “OK”. Both the 2D and 3D views will navigate to put that point at the center of the view without changing the zoom level.
Details:
- the values should be in microns
- you can paste in
x, y, zcoordinates or justx, y; in the latter case, the currentzvalue will not change - any brackets or commas will be removed
- thus you can copy and paste from Horta, or from eg Matlab or Python
Again, HortaCloud users should see “AppStream Basics” for how to move data from your local system to the AppStream Clipboard.
3 - Tracing neurons
In this tutorial, we’ll go over the steps needed to begin tracing a neuron in Horta in the 3D view.
HortaCloud-focused
This tutorial is focused on HortaCloud. However, most of it applies to the desktop version of Horta as well. If you can launch Horta on the desktop, and you see data in the Data Explorer, you should be able to follow most of the other steps of the tutorial.3D-focused
This tutorial doesn’t cover tracing in 2D in detail. Most of it is the same (creating workspaces, neurons, shift-click to add points), but there are some slight differences. See the “Basic Operations” section for details on 2D tracing.
For HortaCloud, 2D tracing is rarely useful, as HortaCloud does not usually contain high-resolution 2D data. You’re not going to be able to accurately place annotations with the low-resolution data.
Prerequisites
- You should be able to log into HortaCloud and launch the Horta application, as detailed in the first tutorial.
- You should be able to open a sample in 3D and navigate the data, as detailed in the second tutorial.
- Your HortaCloud instance should have access to the Janelia MouseLight data on Amazon AWS S3.
- If not, you should have access to some similar data in your HortaCloud instance.
Steps
Start the Horta Application and open a sample in Horta
As detailed in the previous two tutorials:
- Launch HortaCloud
- Locate the
2018-07-02sample in the Data Explorer and open it in the Horta Control Center - Open the sample in the 3D viewer
Create a workspace
The sample that is listed in the Data Explorer, that we’ve opened in Horta, is the representation of the image data in the database. In general, a user never needs to change this information–it’s a static snapshot of the image data. The sample is shared among all users who will work with that dataset.
A workspace, by contrast, contains the neurons that a user has traced. The workspace initially belongs to and can only be seen by the user that created it, but it can then be shared with other users and/or groups.
To create a workspace:
- Open a sample in Horta (as done in the previous step); this is the dataset that the neuron tracing will be associated with
- In the Horta Control Center at right, near the top, just under the “Open 2d” checkbox, click the button with the “+” on it.
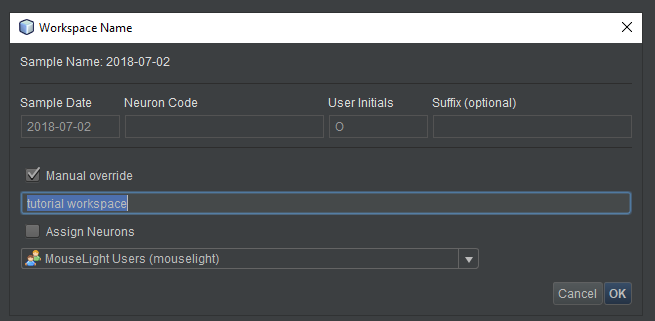
- By default, the dialog box that pops up provides a template for naming the workspace. For this tutorial, however, click the “Manual override” button to name the workspace without using the template. You can use the default name “new workspace” or enter another name, perhaps “tutorial workspace”. For this tutorial, leave “Assign neurons” unchecked.
- Click “OK”
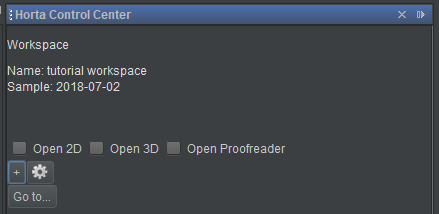
The system will work for a moment, then it will reload the images, this time loading the workspace instead of the sample. You will notice that at the top of the Horta Control Center, the name you gave the workspace will appear just above the sample name now.
In the Data Explorer, this workspace will be located in your “Home” folder (the one with your username next to it in gold), within the “Workspaces” subfolder. You may need to click the refresh button (upper left corner, two arrows in a circle) in the Data Explorer before it shows up.
By default, you are the only person who can see, open, or edit the workspace, save Horta administrators. See the “Basic Operations” section for how to share data.
Adjust colors and save the color model
One immediate advantage of the workspace is the ability to save color settings, which Horta calls the “color model”. Unlike the sample, which is (potentially) shared among many or all users, workspaces often belong to one user. They are therefore appropriate for storing information like color settings, which are often a matter of personal preference.
- First, if the 3D view is not open, click the “Open 3D” checkbox and/or switch to the “Horta 3D” tab as needed.
- Then adjust colors as described in the previous tutorial and shown in screenshots therein. For this sample: set the second purple slider to midway between the left edge and the center lock icons; set the first green slider a bit to the left of the second slider; and hide the third blue channel.
- Next, to save the color model, locate the gear menu in the 3D “Color Sliders” panel, to the right, just under the three channels’ color swatches. Note that the “Horta 2D” tab has its own color sliders. Be sure to use the 3D sliders for this tutorial.
- From the gear menu, choose “Save Color Model to Workspace”.
When you save a color model, all of the color slider settings are saved: channel colors, channel visibility, all sliders, and all locks. From this point forward, when you open this workspace, the color model will be loaded. If you change the color settings later and want to keep them, you’ll need to save them again, however. The color model does not automatically save itself when changed.
The same applies to 2D; you can save the 2D color model (separately) from the gear menu in the 2D color slider area in the Horta 2D tab.
Create a neuron
Now, deep in the third tutorial, we are finally ready to trace a neuron!
In Horta, tracing neurons is done by placing a series of connected point annotations along the neuron signal in the images. These points form one or more trees of points, with each point having one parent point and zero, one, or more child points.
The Horta Control Center contains, below the “Workspace” area, a list of neurons and controls for interacting with neurons.
To create a neuron, click the “+” button below the neuron list, just below the “Spatial filter” label. The default name is “Neuron 1”, where the number will be incremented to be larger than existing neurons. You may also give the neuron any name you like, with some restrictions (ie, the * and ? characters can’t be used). You can rename the neuron at any time by right-clicking its name in the neuron list and choosing “Rename”. Once the neuron is created, the name will appear in the neuron list, and the neuron will be selected (highlighted).
The initial color is randomly chosen from a palette of about twenty. If you’d like to change the color of the neuron, click the color swatch to the right of the neuron’s name in the list and choose a new color from the dialog box.
Add points
Let’s find a neuron to trace. You can find any neuron you like (especially if you are not using the 2018-07-02 sample), but we’ll provide the location of a sample neuron if you want to follow along closely. This neuron is chosen for demonstration purposes only! It may not even be traceable along its entire length.
To find the sample neuron:
- Right-click in the 3D view and choose “Reset Horta Rotation” so you are oriented in the default direction
- Click the “Go to…” button in the Horta Control Center (described in more detail in the previous tutorial)
- Enter the following coordinates and click OK:
[74130,18910,35035]- Remember, if you copy and paste these coordinates, you’ll need to choose the middle icon on the AppStream toolbar and choose “Past to remote session” to transfer the coordinates from your local computer’s clipboard to the AppStream clipboard; once it’s there, you can use control-V to paste as usual
- See “AppStream Basics” for more information on copying and pasting to and from AppStream
We’re going to trace a small part of the bright neuron making a hairpin turn from the right edge of the screen.
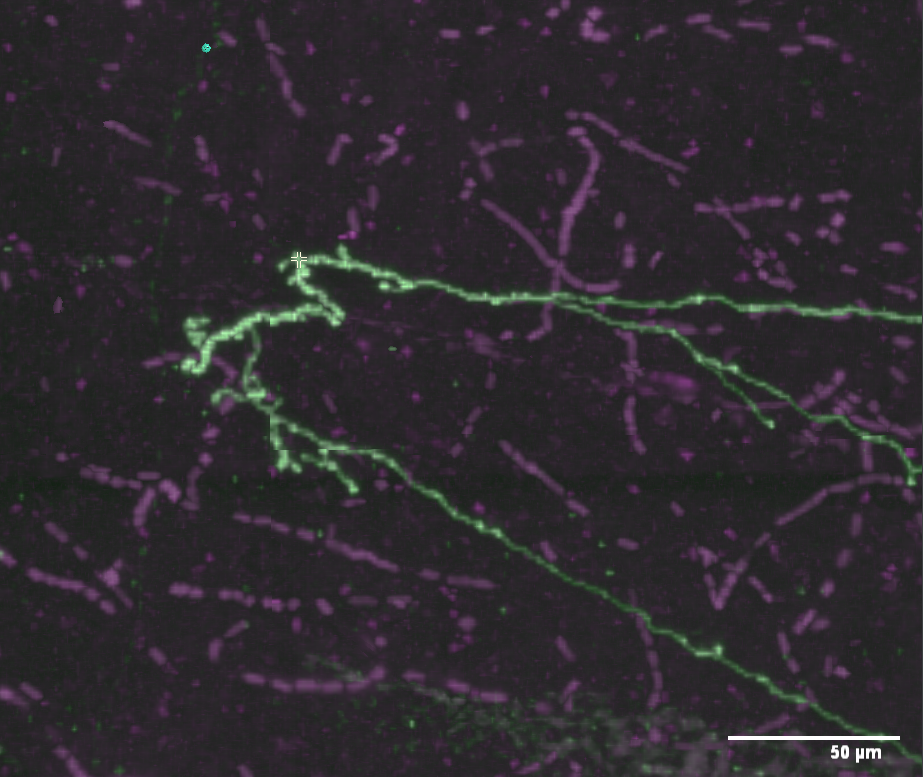
The neuron you want to trace (“Neuron 1” in our case) should be highlighted in the neuron list. If it isn’t, single click it. The list of annotations below the neuron list should be empty at this time, if you’re following the tutorial exactly.
Navigate to the place you want to start tracing. Often this will be the soma of the neuron, but it can be anywhere. For this tutorial, we’ll start placing points in the middle of it. By single-clicking to recenter, dragging the image, and zooming in and out, center the neuron in the view. Feel free to rotate if you like, but to make this tutorial easy to follow from the screenshots, we’ll keep the default rotation.
Now move the mouse pointer over the signal, and over places near the signal but not on it. You’ll notice that a blob with the letter “P” in it appears and reappears as you move the mouse. In fact, you will see that it snaps to the brightest pixels in a small neighborhood around the mouse pointer. If there are no bright pixels, it may not appear at all. This “P” cursor indicates where the point will be placed. Zoom in as much as you need to place the point accurately. Shift-left-click to place the first point when the “P” blob is visible and on top of signal. Near the (x, y, z) location shown above is a good place to place the first point.
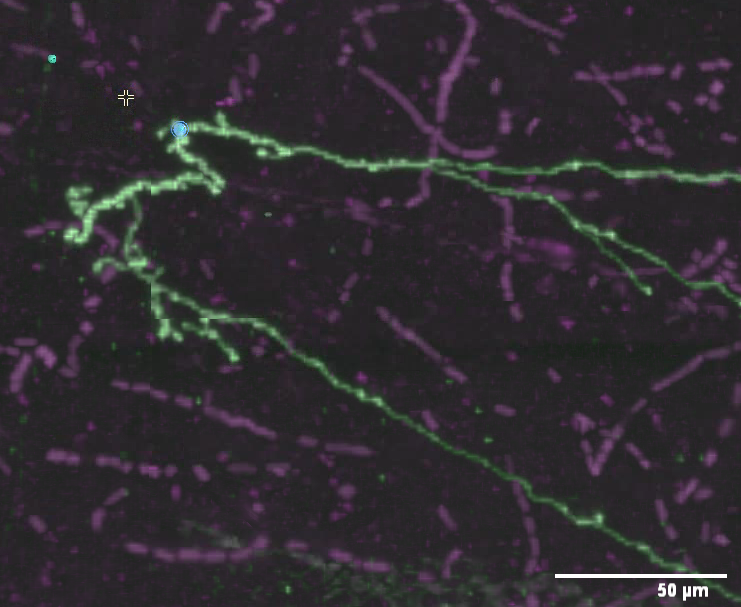
The first point appears! It’ll be drawn in the color of the neuron, and it will also have a “P” indicator drawn on it (although that indicator may be difficult to read depending on the strength of the signal nearby). At this point, if you were to rotate the view, you’ll see that the point lies properly on top of the signal in all dimensions. This is another feature of the “P” blob cursor: it senses the correct depth at which to place points. That is, it not only finds bright pixels in the plane of the display, it also finds the brightest pixel along the axis perpendicular to the screen (with a larger search neighborhood).
Move the mouse cursor along the neuron signal and shift-click again, working to the right of the screen on the upper segment. Repeat a few times until you have a short string of annotations, points connected by lines. The most recent point you’ve placed always has a (sometimes faint) “P” icon. “P” stands for “Parent”: that is the point to which your next point will be connected.
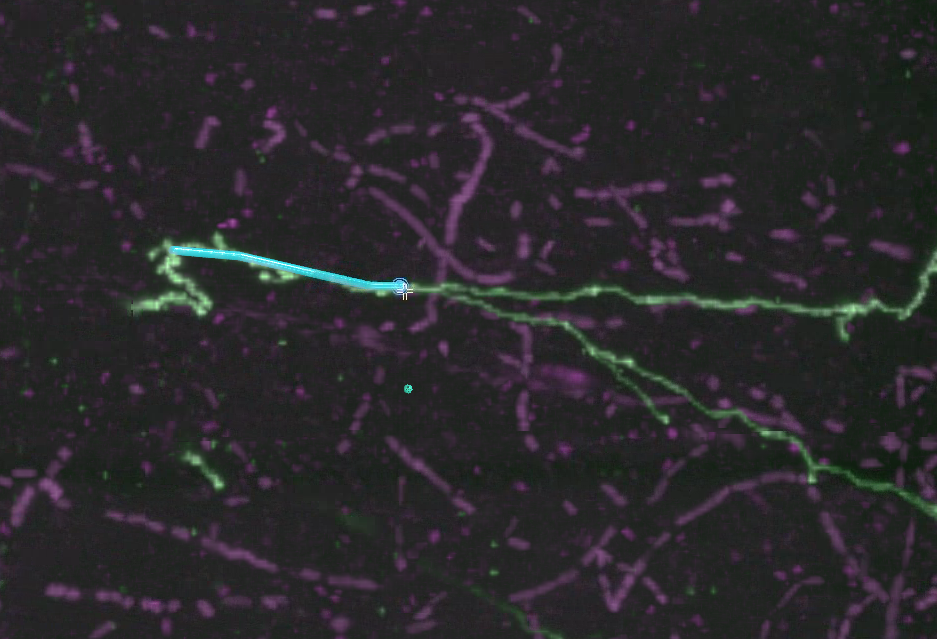
How densely should you place points? That depends on your intended scientific analysis. If you intend to determine region-to-region connectivity, you can annotate quite sparsely. However, if you intend to calculate neuron length or analyze neuron morphology, you will want to place points with smaller separation. If you’re using the tracing as ground truth for machine learning purposes, you may want near pixel-perfect tracing. In this case, look at the main documentation for the “automatically traced path” feature, in which you can have the computer trace paths between human placed points.
The annotation list and more navigation
The annotations list now contains a summary of the points you’ve placed. Not every point is listed–that would quickly get unwieldy–but the “interesting” points are. By default, the list includes every root point, endpoint, and branch point. It does not include points along a linear chain. But it also includes points that have some kind of user annotation attached to them (discussed elsewhere in the documentation). If you’re following closely, there will only be two points in the annotation list at this time, the root point and the endpoint.
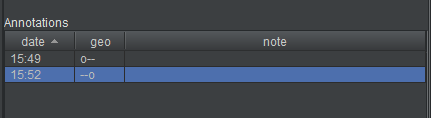
Both the annotation list and the neuron list can be used for navigation.
- if you double-click a neuron in the neuron list, the view will center on the first point (the root point) in the neuron
- if you double-click on an annotation in the annotation list, the view will center on it
Moving and deleting points
If you make any errors when adding points, there are easy ways to correct them.
If you need to move a point a short distance, left-click and drag it.
- in 2D, you can only drag in the same plane
- in 3D, you can again only drag in the plane of the view; the point does not snap to bright pixels when you drag, so you’ll probably have to rotate and drag multiple times from different angles to fine-tune the location
To delete a point:
- in 2D: left click on it, then press the delete key on the keyboard
- in 2D: right-click on it, then choose “Delete link” from the menu
- in 3D: right-click on it, then choose “Delete Vertex” from the menu
If you delete an annotation in a linear chain, it’ll connect the two surrounding points. You can’t delete the root of the tree or branch points (see below for more on branching). If you need to delete multiple points, see the “Delete subtree” feature described in the “Basic Operations” section.
Branching
You will see that each time you shift-click, the “P” indicator moves to the point you’ve most recently placed. “P” stands for “Parent”: that is the point to which your next point will be connected. This is how you’ll create branches.
If you’ve been following this tutorial closely, your neuron should be approaching a place where the signal branches. Continue adding points headed left until you reach the branch, and place a point directly on the branch location. If you’re tracing on different data, find a convenient branch point, or imagine one (it’s only a tutorial!).
At this point, the next parent icon (“P”) should be on the branch. Choose one arm of the neuron to trace, and add a few points. Now, go back to the branch point and single-click it so it again shows the “P” icon. This indicates that the next point you place will use this point (the branch point) as its parent. Add a few more points along the other branch. You’ve now got a branching structure.
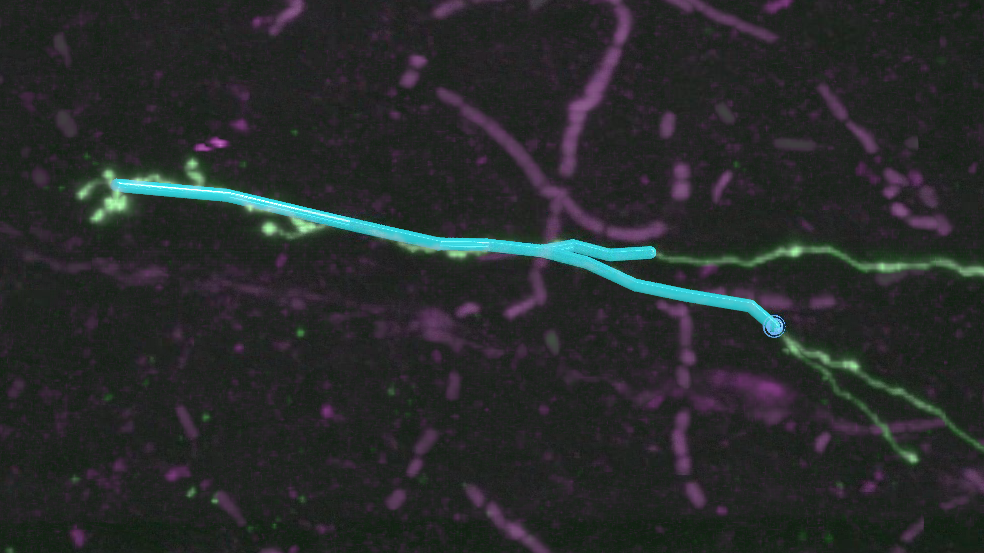
The annotation list now shows the root point, the branch point, and two endpoints.
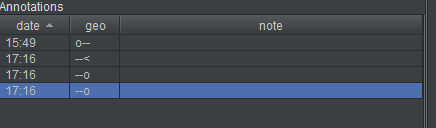
There are workflows that help you manage the task of tracing a branched structure. See, for example, “Simple workflow with notes and filters” in the “Features” section
Continuing work in the workspace
Annotations are saved to the database immediately after they are placed. This goes for all operations; the application is in constant communication with the database, and all changes are saved immediately. You can quit at any time, and no work will be lost. In this respect, HortaCloud acts more like a mobile application than a desktop application.
When you want to continue work, simple open the workspace from the Data Explorer just as you earlier opened a sample. That is, right-click it in the Data Explorer and choose “Open in Horta”. The neuron data will load immediately. After that, you can choose “Open 3D” and continue work tracing.
4 - Exporting and importing neurons
In this tutorial, we’ll go over how to export and import traced neurons from/to Horta.
Prerequisites
- You should be able to log into HortaCloud and launch the Horta application, as detailed in the first tutorial.
- You should be able to open a workspace in 3D and navigate the data, as detailed in the second tutorial.
- You should be able to create and add points to a neuron, as detailed in the third tutorial.
Steps
Open the workspace
Launch HortaCloud and open the workspace created in the previous tutorial, or any other workspace that you can trace in.
Exporting neurons as SWC files
Once you’ve finished tracing a neuron, you will probably want to export the data for analysis or visualization in another piece of software. Horta supports import and export to the standard SWC file format. Details of the SWC format can be found in the “Reference” section.
To export a traced neuron:
- Right click on its name in the neuron list and choose “Export SWC File”. A file dialog box will open.
- Name the file whatever you like. An extension of “.swc” is strongly recommended. The two options on the right are discussed in the “Basic Operations” section. The defaults are appropriate almost all of the time.
- Choose the export location. For desktop, choose any location you like on your hard drive. For HortaCloud, we need to save to an intermediate location first. At the top of the dialog box, click the “Save in” drop-down menu and choose “Temporary Files”.
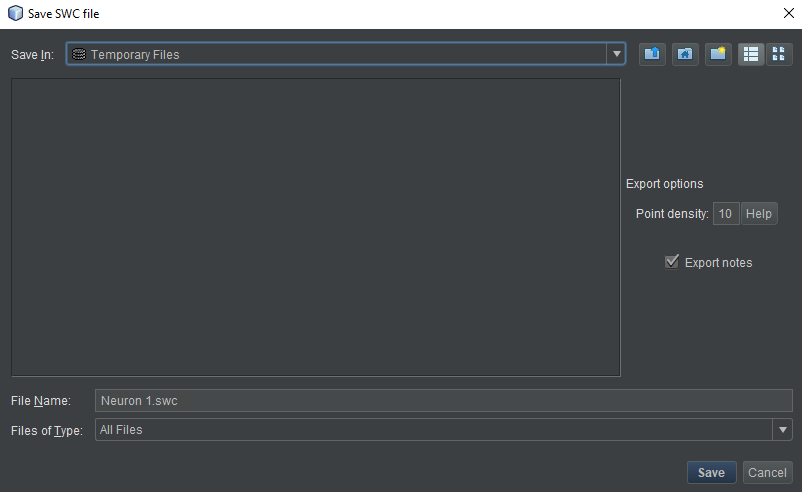
- Click “OK”. A “Background Tasks” window will open with a progress bar. This operation is very fast, and you may not even see the bar before it’s filled. For desktop Horta, you’re done after this step.
- For HortaCloud users, you now need to download your file(s) from the “Temporary Files” location on AppStream. The AppStream toolbar appears at the top of the browser’s content window, above the top of the Horta main window. Click the third icon from left, which has a tooltip of “My Files”.
- In the new dialog box that opens, click “Temporary Files”. This is a web view into the folder where you exported your neuron. At the right end of the row for each file in this directory is a downward facing chevron. Click it and choose “Download” from the drop-down menu. Depending on your browser, you may be prompted to choose a location, or the file may be downloaded to your default downloads location. Also depending on your browser, you may need to authorize or confirm the download from the AppStream site.
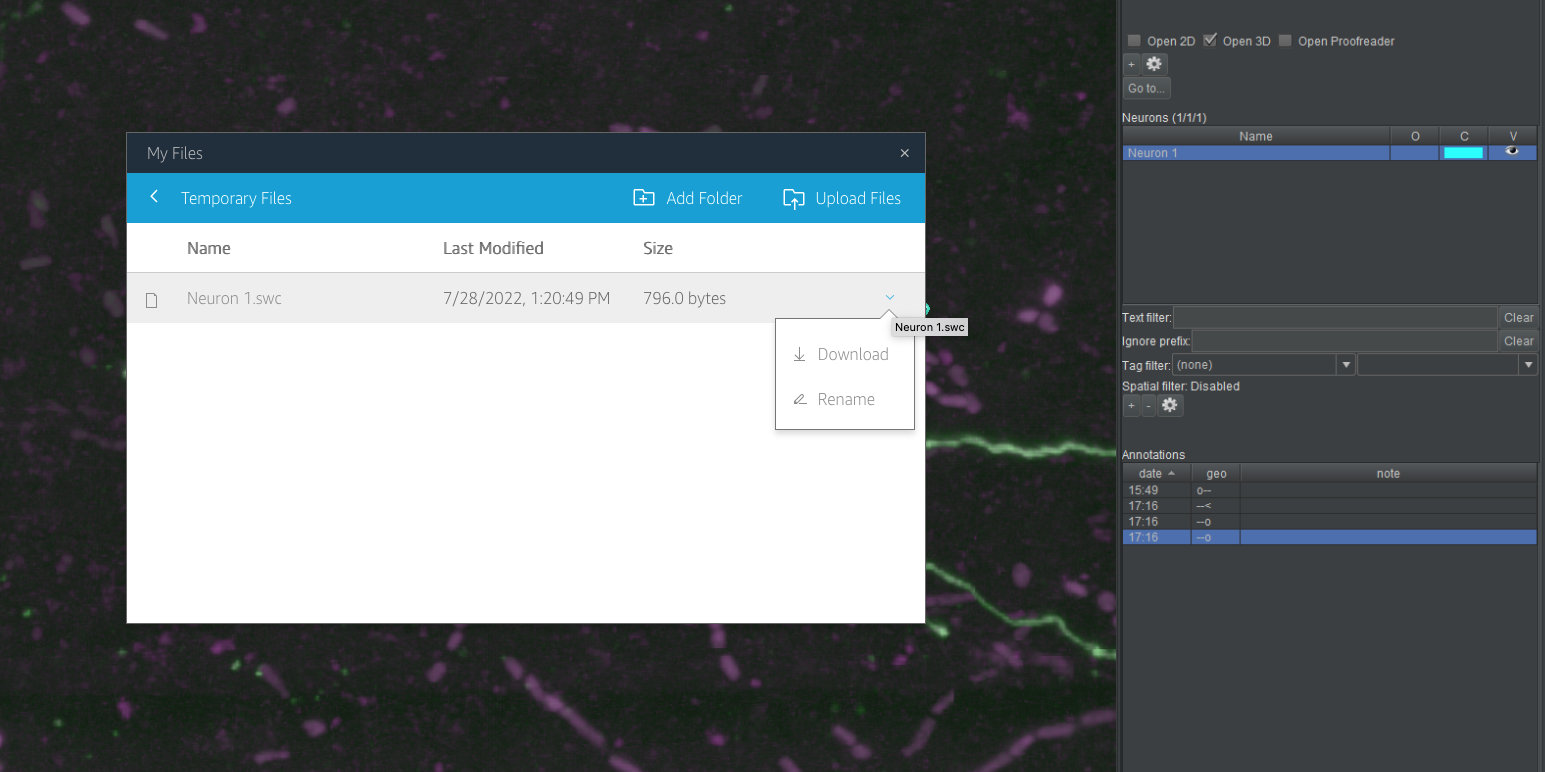
Safari
File downloads seem not to work in Safari. Use Firefox or Chrome on Mac if you need to download from “Temporary Files”. (Strangely, uploading files to the same location does work in Safari.)Temporary means temporary!
The name “Temporary Files” is accurate! The files in this location will be deleted when your session ends. You must download any exported neurons to your local computer in the same session in which you exported them.Importing neurons from SWC files
Importing neurons from SWC files is basically the same as exporting, in reverse. Again, the HortaCloud procedure involves an intermediate location.
If you’ve been using the Janelia 2021-03-17 sample for your test, you can download a test neuron to import (right-click and download the linked file). If you don’t have access to the Janelia sample we’re using in these tutorials, you can test importing a neuron into your own sample and workspace by simply exporting any neuron (as in the previous section), deleting the neuron using the “-” button under the neuron list, and then reimporting the same neuron.
To import a traced neuron:
- Open a workspace that corresponds to the same sample the neuron was exported from. In other words, the exported neuron’s SWC file must be using the same coordinate system and same scale as the target sample. If not, the imported neuron will not lie on top of the neuron signal in the images. In many cases, it will be imported somewhere in space far from the brain imagery.
- (HortaCloud only) Upload the SWC file to AppStream. Click the “My Files” icon on the AppStream toolbar (third from left). Click “Temporary Files”. You can drag files directly to this dialog box, and they will be uploaded. Alternately, click “Upload files” and choose the files from the file dialog that opens.
- In Horta, click the gear menu under the workspace information area, above the neuron list. Choose “Import SWC file as separate neurons”.
- Desktop users may choose any neuron on their hard disk at this point. HortaCloud users should again navigate to the “Temporary Files” area by using the drop-down menu called “Look in” at the top. Either way, choose a neuron SWC file and then click “Open”.
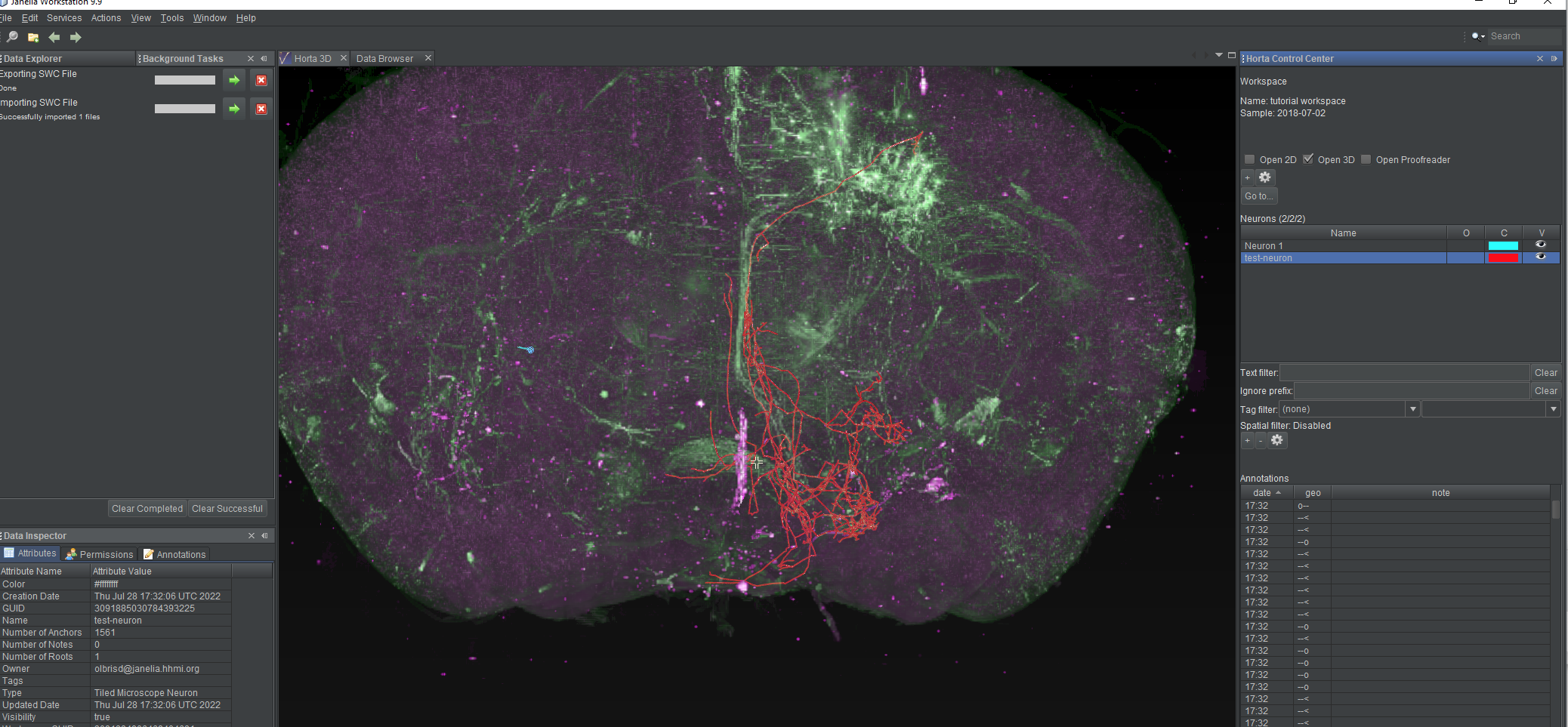
- The “Background Tasks” window will again open, and the neuron will appear in the neurons list, and in the 2D or 3D views, if you have them open.
Once the neuron has loaded, it will be visible in the 3D view, the 2D view (if it’s open), and the neuron list. If you select the test neuron in the neuron list, you’ll see a large number of branch points and endpoints in the annotation list.
Bulk import/export
See the “Basic Operations” section for more information on exporting multiple neurons at once, and on importing multiple neurons either interactive or in a offline batch process.
What’s next
After finishing these tutorials, you may want to read the “Basic Operations” section of the User Manual. This section will cover much of the same material discussed in these tutorials, with a few more details and a few more operations described. The “Features” section lists many less-commonly used tools. The “Reference” section contains details on, for example, file formats. Typically users will rarely need to consult that section.