Overview
HortaCloud is a streaming 3D annotation platform for large microscopy data that runs entirely in the cloud. It is a free, open source research software tool, developed by Janelia Research Campus.
It combines state-of-the-art volumetric visualization, advanced features for 3D neuronal annotation, and real-time multi-user collaboration with a set of enterprise-grade backend microservices for moving and processing large amounts of data rapidly and securely. HortaCloud takes advantage of cloud-based Virtual Desktop Infrastructure (VDI) to perform all 3D rendering in cloud-leased GPUs which are data-adjacent, and only transfer a high-fidelity interactive video stream to each annotator’s local compute platform through a web browser.
What is it good for?: HortaCloud is a powerful tool for 3D visualization and annotation of large-scale microscopy data. It has been used to trace axons across the entire mouse brain by Janelia’s MouseLight Team Project.
What is it not good for?: HortaCloud is a very specialized tool for sparse microscopy data. It is not intended for annotation of dense data sets, such as electron microscopy (EM) imagery.
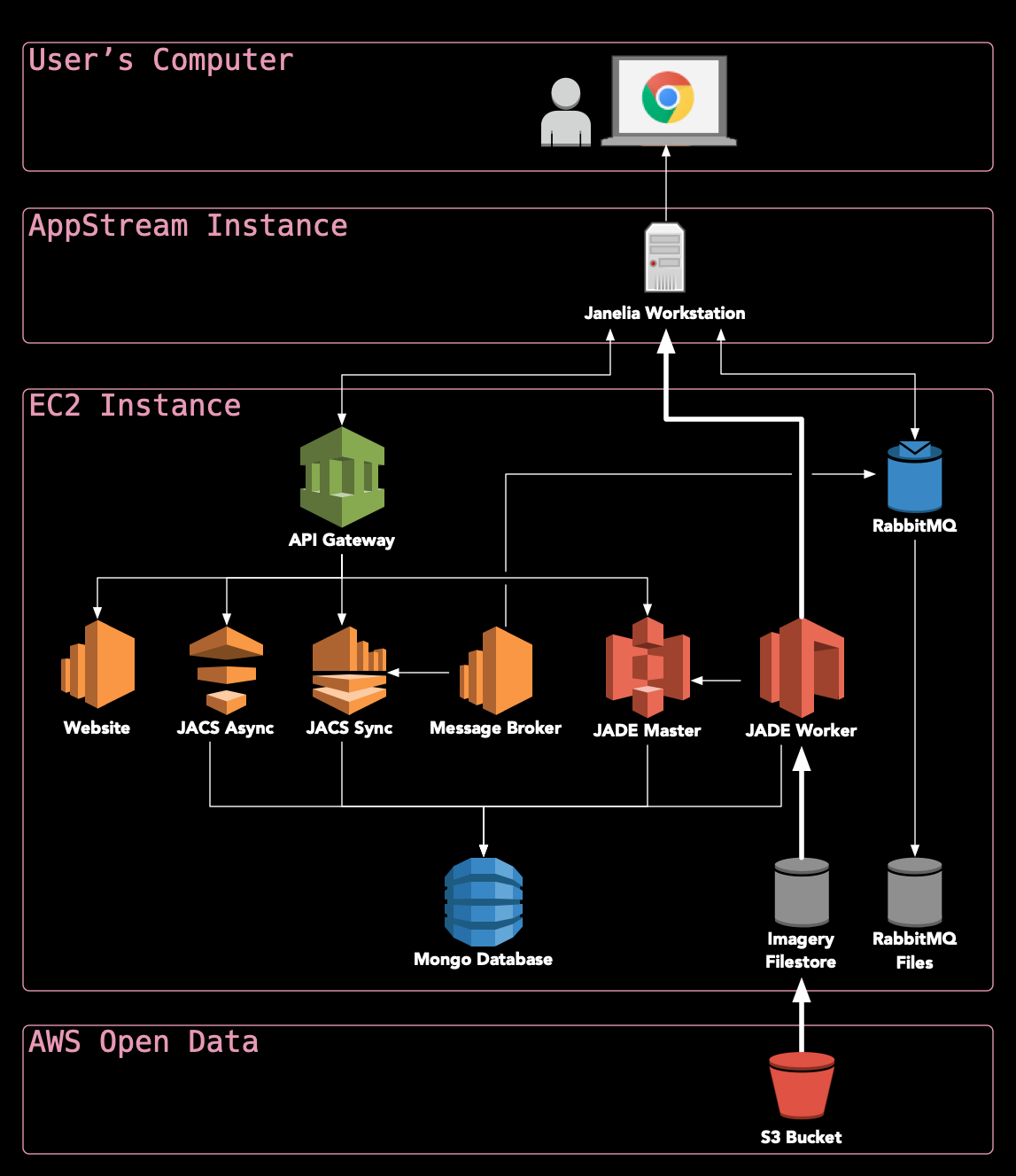
What makes HortaCloud unique?: Leveraging state-of-the-art services on AWS allows HortaCloud to run entirely in the cloud, making it possible to visualize terabyte-scale 3D volumes without moving all of that data over the Internet (see diagram below).
Where should I go next?
If you are a HortaCloud user, read through the User Manual to get familiar with the tools.
If you are a system administrator or developer looking to deploy an instance of HortaCloud, start with the AWS Deployment Guide.
Feedback
Was this page helpful?
Glad to hear it! Please tell us how we can improve.
Sorry to hear that. Please tell us how we can improve.